Usage réservé à la recherche
p47phox Antibody [L8D20]
N° de cat.: F4804
Application:
Réactivité:
Informations dutilisation
| Dilution |
|---|
|
| Application |
|---|
| WB, IP, IHC |
| Réactivité |
|---|
| Human |
| Source |
|---|
| Rabbit Monoclonal Antibody |
| Tampon de stockage |
|---|
| PBS, pH 7.2+50% Glycerol+0.05% BSA+0.01% NaN3 |
| Stockage (à partir de la date de réception) |
|---|
| -20°C (avoid freeze-thaw cycles), 2 years |
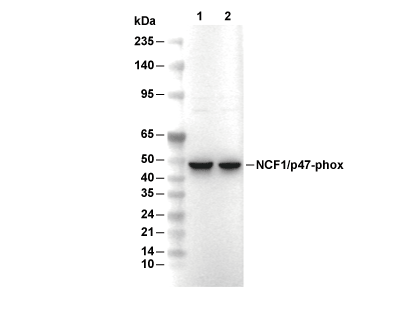
| Poids moléculaire prédit Poids moléculaire observé |
|---|
| 44 kDa 45 kDa |
| *Pourquoi les poids moléculaires prédit et réel diffèrent-ils? Les raisons suivantes peuvent expliquer les différences entre le poids moléculaire prédit et réel des protéines. |
| Contrôle positif | Human spleen tissue; Raji cells; Ramos cells |
|---|---|
| Contrôle négatif |
Méthodes expérimentales
| WB |
|---|
Experimental Protocol:
Sample preparation
1. Tissue: Lyse the tissue sample by adding an appropriate volume of ice-cold RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail),and homogenize the tissue at a low temperature. 2. Adherent cell: Aspirate the culture medium and wash the cells with ice-cold PBS twice. Lyse the cells by adding an appropriate volume of RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail) and put the sample on ice for 5 min. 3. Suspension cell: Transfer the culture medium to a pre-cooled centrifuge tube. Centrifuge and aspirate the supernatant. Wash the cells with ice-cold PBS twice. Lyse the cells by adding an appropriate volume of RIPA/NP-40 Lysis Buffer (containing Protease Inhibitor Cocktail) and put the sample on ice for 5 min. 4. Place the lysate into a pre-cooled microcentrifuge tube. Centrifuge at 4°C for 15 min. Collect the supernatant;
5. Remove a small volume of lysate to determine the protein concentration;
6. Combine the lysate with protein loading buffer. Boil 20 µL sample under 95-100°C for 5 min. Centrifuge for 5 min after cool down on ice.
Electrophoretic separation
1. According to the concentration of extracted protein, load appropriate amount of protein sample and marker onto SDS-PAGE gels for electrophoresis. Recommended separating gel (lower gel) concentration: 10%. Reference Table for Selecting SDS-PAGE Separation Gel Concentrations 2. Power up 80V for 30 minutes. Then the power supply is adjusted (110 V~150 V), the Marker is observed, and the electrophoresis can be stopped when the indicator band of the predyed protein Marker where the protein is located is properly separated. (Note that the current should not be too large when electrophoresis, too large current (more than 150 mA) will cause the temperature to rise, affecting the result of running glue. If high currents cannot be avoided, an ice bath can be used to cool the bath.)
Transfer membrane
1. Take out the converter, soak the clip and consumables in the pre-cooled converter;
2. Activate PVDF membrane with methanol for 1 min and rinse with transfer buffer;
3. Install it in the order of "black edge of clip - sponge - filter paper - filter paper - glue -PVDF membrane - filter paper - filter paper - sponge - white edge of clip"; 4. The protein was electrotransferred to PVDF membrane. ( 0.45 µm PVDF membrane is recommended ) Reference Table for Selecting PVDF Membrane Pore Size Specifications Recommended conditions for wet transfer: 200 mA, 120 min. ( Note that the transfer conditions can be adjusted according to the protein size. For high-molecular-weight proteins, a higher current and longer transfer time are recommended. However, ensure that the transfer tank remains at a low temperature to prevent gel melting.)
Block
1. After electrotransfer, wash the film with TBST at room temperature for 5 minutes;
2. Incubate the film in the blocking solution for 1 hour at room temperature;
3. Wash the film with TBST for 3 times, 5 minutes each time.
Antibody incubation
1. Use 5% skim milk powder to prepare the primary antibody working liquid (recommended dilution ratio for primary antibody 1:1000), gently shake and incubate with the film at 4°C overnight; 2. Wash the film with TBST 3 times, 5 minutes each time;
3. Add the secondary antibody to the blocking solution and incubate with the film gently at room temperature for 1 hour;
4. After incubation, wash the film with TBST 3 times for 5 minutes each time.
Antibody staining
1. Add the prepared ECL luminescent substrate (or select other color developing substrate according to the second antibody) and mix evenly;
2. Incubate with the film for 1 minute, remove excess substrate (keep the film moist), wrap with plastic film, and expose in the imaging system. |
| IHC |
|---|
Experimental Protocol:
Deparaffinization/Rehydration
1. Deparaffinize/hydrate sections:
2. Incubate sections in three washes of xylene for 5 min each.
3. Incubate sections in two washes of 100% ethanol for 10 min each.
4. Incubate sections in two washes of 95% ethanol for 10 min each.
5. Wash sections two times in dH2O for 5 min each.
6.Antigen retrieval: For Citrate: Heat slides in a microwave submersed in 1X citrate unmasking solution until boiling is initiated; continue with 10 min at a sub-boiling temperature (95°-98°C). Cool slides on bench top for 30 min.
Staining
1. Wash sections in dH2O three times for 5 min each.
2. Incubate sections in 3% hydrogen peroxide for 10 min.
3. Wash sections in dH2O two times for 5 min each.
4. Wash sections in wash buffer for 5 min.
5. Block each section with 100–400 µl of blocking solution for 1 hr at room temperature.
6. Remove blocking solution and add 100–400 µl primary antibody diluent in to each section. Incubate overnight at 4°C.
7. Remove antibody solution and wash sections with wash buffer three times for 5 min each.
8. Cover section with 1–3 drops HRPas needed. Incubate in a humidified chamber for 30 min at room temperature.
9. Wash sections three times with wash buffer for 5 min each.
10. Add DAB Chromogen Concentrate to DAB Diluent and mix well before use.
11. Apply 100–400 µl DAB to each section and monitor closely. 1–10 min generally provides an acceptable staining intensity.
12. Immerse slides in dH2O.
13. If desired, counterstain sections with hematoxylin.
14. Wash sections in dH2O two times for 5 min each.
15. Dehydrate sections: Incubate sections in 95% ethanol two times for 10 sec each; Repeat in 100% ethanol, incubating sections two times for 10 sec each; Repeat in xylene, incubating sections two times for 10 sec each.
16. Mount sections with coverslips and mounting medium.
|
Description biologique
| Spécificité |
|---|
| p47phox Antibody [L8D20] detects endogenous levels of total p47phox protein. |
| Localisation subcellulaire |
|---|
| Cytoplasm, Membrane |
| Uniprot ID |
|---|
| P14598 |
| Clone |
|---|
| L8D20 |
| Synonyme |
|---|
| NOXO2; SH3PXD1A; NCF1; Neutrophil cytosol factor 1; NCF-1; 47 kDa neutrophil oxidase factor; NCF-47K; Neutrophil NADPH oxidase factor 1; Nox organizer 2; Nox-organizing protein 2; SH3 and PX domain-containing protein 1A; p47-phox |
| Contexte |
|---|
| p47phox (neutrophil cytosolic factor 1, NCF1) is a critical organizer subunit of the phagocyte NADPH oxidase 2 (NOX2) complex, which is essential for innate immunity by coordinating the assembly of cytosolic factors (p47phox, p67phox, p40phox, Rac) with the membrane-bound cytochrome b558 (gp91phox/NOX2-p22phox) to facilitate superoxide production for microbial killing; its deficiency leads to chronic granulomatous disease (CGD), characterized by recurrent infections. The 390-amino-acid p47phox protein adopts an autoinhibited conformation, featuring an N-terminal phox homology (PX) domain that binds PI(3,4)P2 or phosphatidic acid for membrane targeting, tandem Src homology 3 (SH3) domains that, in the resting state, interact with an autoinhibitory region (AIR) to block activity, a C-terminal proline-rich region (PRR), and a serine-rich tail (Ser303–Ser379) containing multiple phosphorylation sites as well as polybasic motifs for initial p22phox docking. Upon pathogen recognition, rapid and multi-site phosphorylation by kinases such as PKC, MAPK, and PAK at C-terminal serines (notably Ser303/304/328/359/370) disrupts the AIR-SH3 interaction, exposes cryptic SH3 domains to engage the p22phox PRR for membrane translocation, releases the PX domain for lipid docking, and recruits p67phox and Rac to activate the flavocytochrome for electron transfer from NADPH to O₂, producing superoxide via ping-pong kinetics. This phosphorylation-driven conformational switch ensures spatial and temporal control of ROS generation at phagosomes, with mutations like Δ219–222 or W193R in p47phox impairing SH3-p22phox binding and reducing superoxide output by 60–100%. Post-translational modifications, such as Tyr159/240 phosphorylation, further enhance oxidase priming. Dysregulated p47phox activity contributes to vascular oxidative stress in hypertension and heart failure through endothelial NOX activation, promotes atherosclerosis via ROS generated by vascular smooth muscle cells and monocytes, and is implicated in inflammatory diseases where NOX2 hyperactivity leads to tissue damage. |
| Références |
|---|
|
Support technique
Tel: +1-832-582-8158 Ext:3
Si vous avez dautres questions, veuillez laisser un message.